We are excited to share with you results from two Zooniverse projects, ‘Etch A Cell – Fat Checker’ and ‘Etch A Cell – Fat Checker Round 2’. Over the course of these two projects, more than 2000 Zooniverse volunteers contributed over 75 thousand annotations!
One of the core aims of these two projects was to enable the design and implementation of machine learning approaches that could automate the annotation of fat droplets in novel data sets, to provide a starting point for other research teams attempting to perform similar tasks.
With this in mind, we have developed multiple machine learning algorithms that can be applied to both 2D and 3D fat droplet data. We describe these models in the blog post below.
| Machine learning model | 2D or 3D data | Publications to date | Other outputs |
| PatchGAN | 2D | https://ceur-ws.org/Vol-3318/short15.pdf | https://github.com/ramanakumars/patchGAN https://pypi.org/project/patchGAN/ |
| TCuP-GAN | 3D | “What to show the volunteers: Selecting Poorly Generalized Data using the TCuPGAN”; Sankar et al., accepted in ECMLPKDD Workshop proceedings. | https://github.com/ramanakumars/TCuPGAN/ |
| UNet/UNet3+/nnUNet | 2D | https://huggingface.co/spaces/umn-msi/fatchecker |
Machine learning models for the segmentation of fat droplets in 2D data
Patch Generative Adversarial Network (PatchGAN)
Generative Adversarial Networks (GANs) were introduced in 2018 for the purpose of realistic learning of image-level features and have been used for various computer vision related applications. We implemented a pixel-to-pixel translator model called PatchGAN, which learns to convert (or “translate”) an input image to another image form. For example, such a framework can learn to convert a gray-scale image to a colored version.
The “Patch” in PatchGAN signifies its capability to learn image features in different sub-portions of an image (rather than just across an entire image as a whole). In the context of the Etch A Cell – Fat Checker project data, predicting the annotation regions of fat droplets is analogous to PatchGAN’s image-to-image translation task.
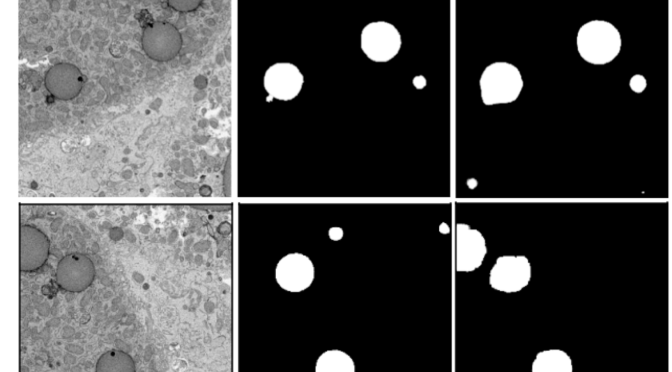
We trained the PatchGAN model framework on the ~50K annotations generated by volunteers in Etch A Cell – Fat Checker. Below we show two example images from the Etch A Cell: Fat Checker (left column) along with aggregated annotations provided by the volunteers (middle panel), and their corresponding 2D machine learning model (PatchGAN) predicted annotations.
We found that the PatchGAN typically performed well in learning the subject image to fat-droplet annotation predictions. However, we noticed that the model highlighted some regions potentially missed by the volunteers, as well as instances where it has underestimated some regions that the volunteers have annotated (usually intermediate to small sized droplets).
We made have made this work, our generalized PatchGAN framework, available via an open-source repository at https://github.com/ramanakumars/patchGAN and https://pypi.org/project/patchGAN/. This will allow anyone to easily train the model on a set of images and corresponding masks, or to use the pre-trained model to infer fat droplet annotations on images they have at hand.
UNet, UNet3+, and nnUNet
In addition to the above-mentioned PatchGAN network, we have also trained three additional frameworks for the task of fat droplet identification – UNet, UNet3+, and nnUNet.
UNet is a popular deep-learning method used for semantic segmentation within images (e.g., identifying cars/traffic in an image) and has been shown to capture intricate image details and precise object delineation. Its architecture is U-shaped with two parts – an encoder that learns to reduce the input image down a compressed “fingerprint” and a decoder which learns to predict the target image (e.g., fat droplets in the image) based on that compressed fingerprint. Fine-grained image information is shared between the encoder and decoder parts using the so-called “skip connections”. UNet3+ is an upgraded framework built upon the foundational UNet that has been shown to capture both local and global features within medical images.
nnUNet is an user-friendly, efficient, and state-of-the-art deep learning platform to train and fine-tune models for diverse medical imaging tasks. It employs a UNet-based architecture and comes with image pre-processing and post-processing techniques.
We trained these three networks on the data from the Fat Checker 1 project. Below, we show three example subject images along with their corresponding volunteer annotations and different model predictions. Between the three models, nnUNet demonstrated superior performance.

Machine learning models for the segmentation of fat droplets in 3D data
Temporal Cubic PatchGAN (TCuP-GAN
Motivated by the 3D volumetric nature of the Etch A Cell – Fat Checker project data, we also developed a new 3D deep learning method that learns to predict the direct 3D regions corresponding to the fat droplets. To develop this, we built on top of our efforts of our PatchGAN framework and merged it with another computer vision concept called “LongShort-Term Memory Networks (LSTMs)”. Briefly, in recent years, LSTMs have seen tremendous success in learning sequential data (e.g., words and their relationship within a sentence) and they have been used to learn relationships among sequences of images (e.g., movement of a dog in a video).
We have successfully implemented and trained our new TCuP-GAN model on the Etch A Cell – Fat Checker data. Below is an example image cube – containing a collection of 2D image slices that you viewed and annotated – along with the fat droplet annotation prediction of our 3D model. For visual guidance, we show the middle panel where we reduced the transparency of the image cube shown in the left panel, highlighting the fat droplet structures that lie within.
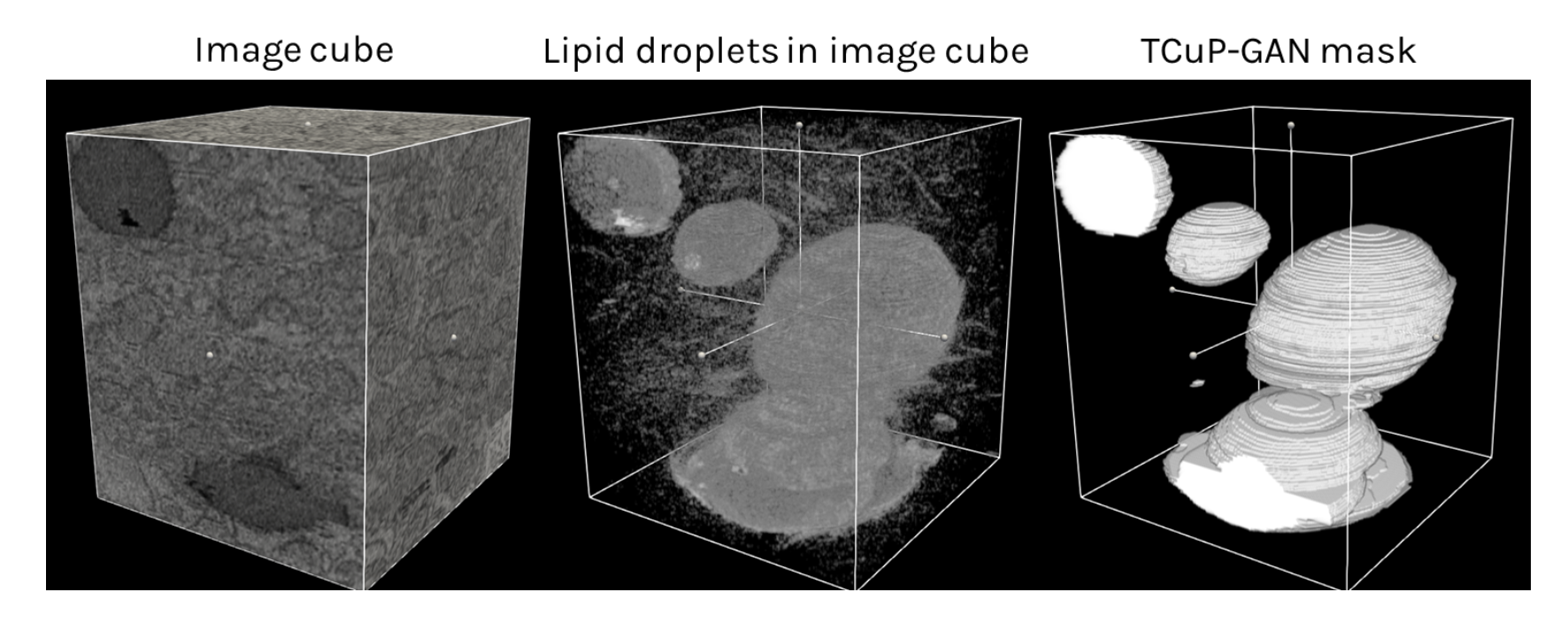
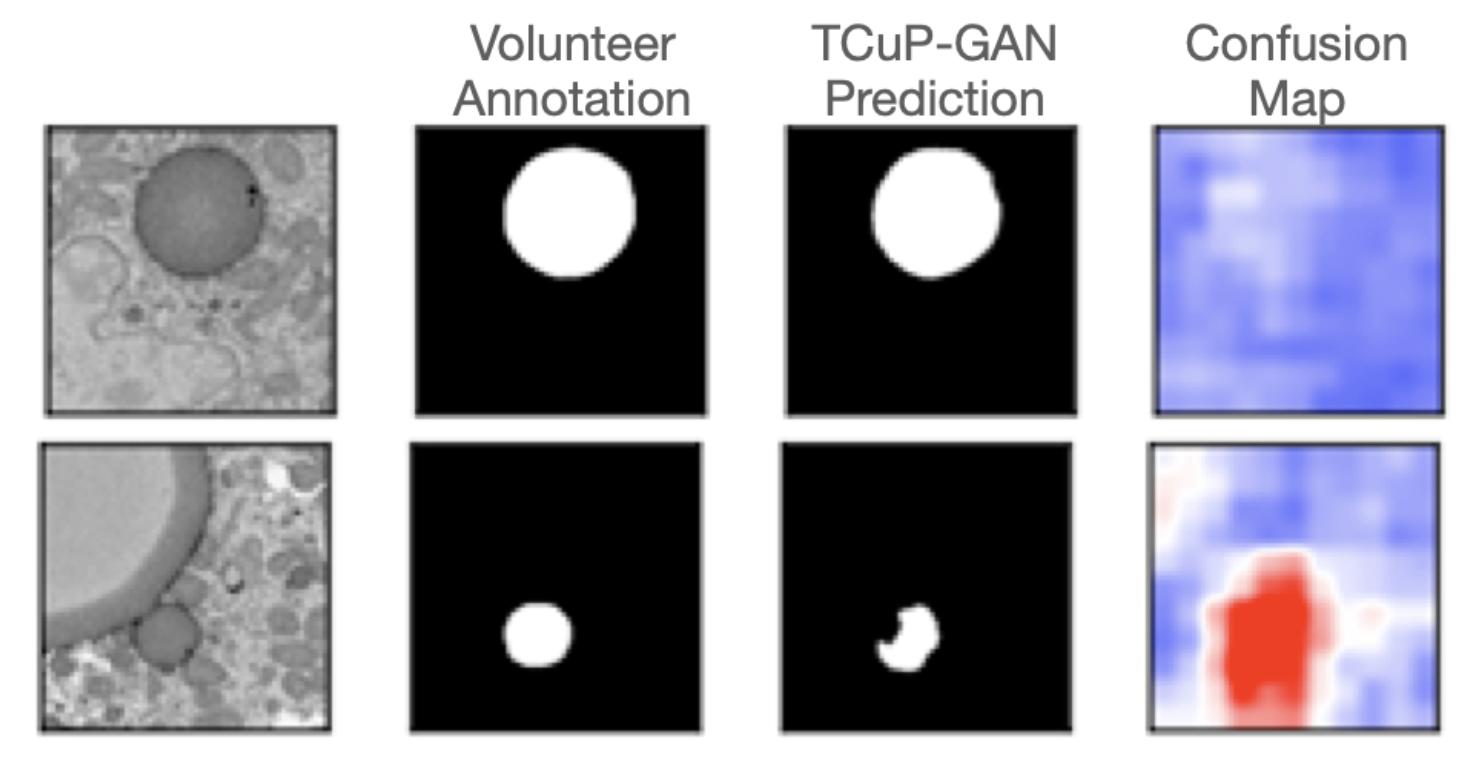
We found that our TCuP-GAN model successfully predicts the 3D fat droplet structures. In doing so, our model also learns realistic (and informative) signatures of lipid droplets within the image. Leveraging this, we are able to ask the model which 2D image slices contain the most confusion between lipid droplets and surrounding regions of the cells when it comes to annotating the fat droplets. Below, we show two example slices where the model was confident about the prediction (i.e., less confusion; top panel) and where the model was significantly confused (red regions in fourth column of the bottom panel). As such, we demonstrated that our model can help find those images in the data set that preferentially require information from the volunteers. This can serve as a potential efficiency step for future research teams to prioritize certain images that require attention from the volunteers.
Integrating Machine Learning Strategies with Citizen Science
Several thousands of annotations collected from the Etch A Cell – Fat Checker project(s) and their use to train various machine learning frameworks have opened up possibilities that can enhance the efficiency and annotation gathering and help accelerate the scientific outcomes for future projects.
While the models we described here all performed reasonably well in learning to predict the fat droplets, there were a substantial number of subjects where they were inaccurate or confused. Emerging “human-in-the-loop” strategies are becoming increasingly useful in these cases – where citizen scientists can help with providing critical information on those subjects that require the most attention. Furthermore, an imperfect machine learning model can provide an initial guess which the citizens can use as a starting point and provide edits, which will greatly greatly reduce the amount of effort needed by individual citizen scientists.
For our next steps, using the data from the Etch A Cell – Fat Checker projects, we are working towards building new infrastructure tools that will enable future projects to leverage both citizen science and machine learning towards solving critical research problems.
Enhancements to the freehand drawing tool
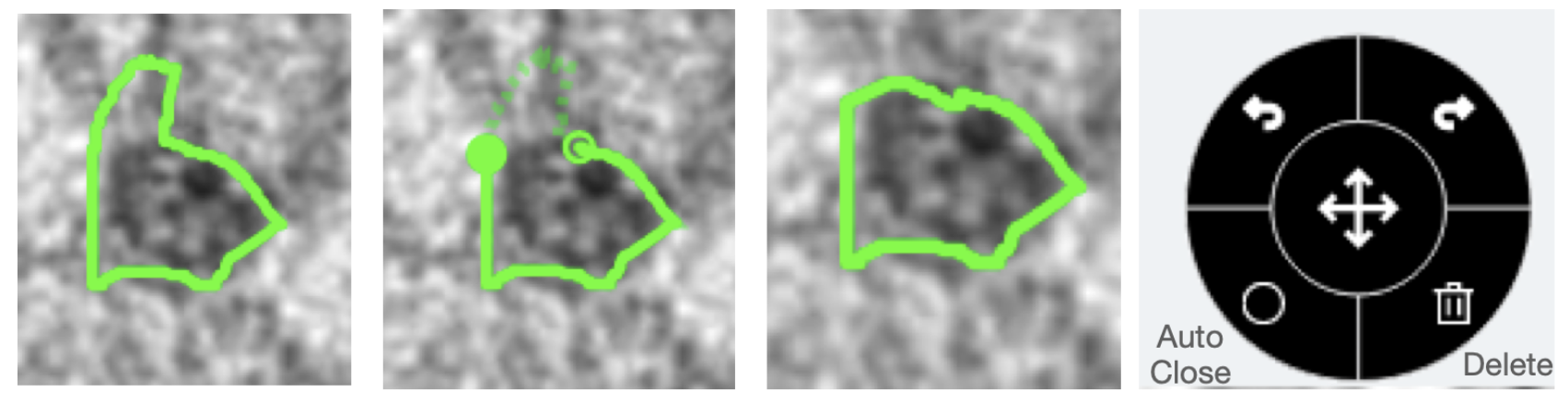
First, we have made upgrades to the existing freehand line drawing tool on Zooniverse. Specifically, users will now be able to edit a drawn shape, undo or redo during any stage of their drawing process, automatically close open shapes, and delete any drawings. Below is a visualization of an example drawing where the middle panel illustrates the editing state (indicated by the open dashed line) and re-drawn/edited shape. The tool box with undo, redo, auto-close, and delete functions is also shown.
A new “correct a machine” framework
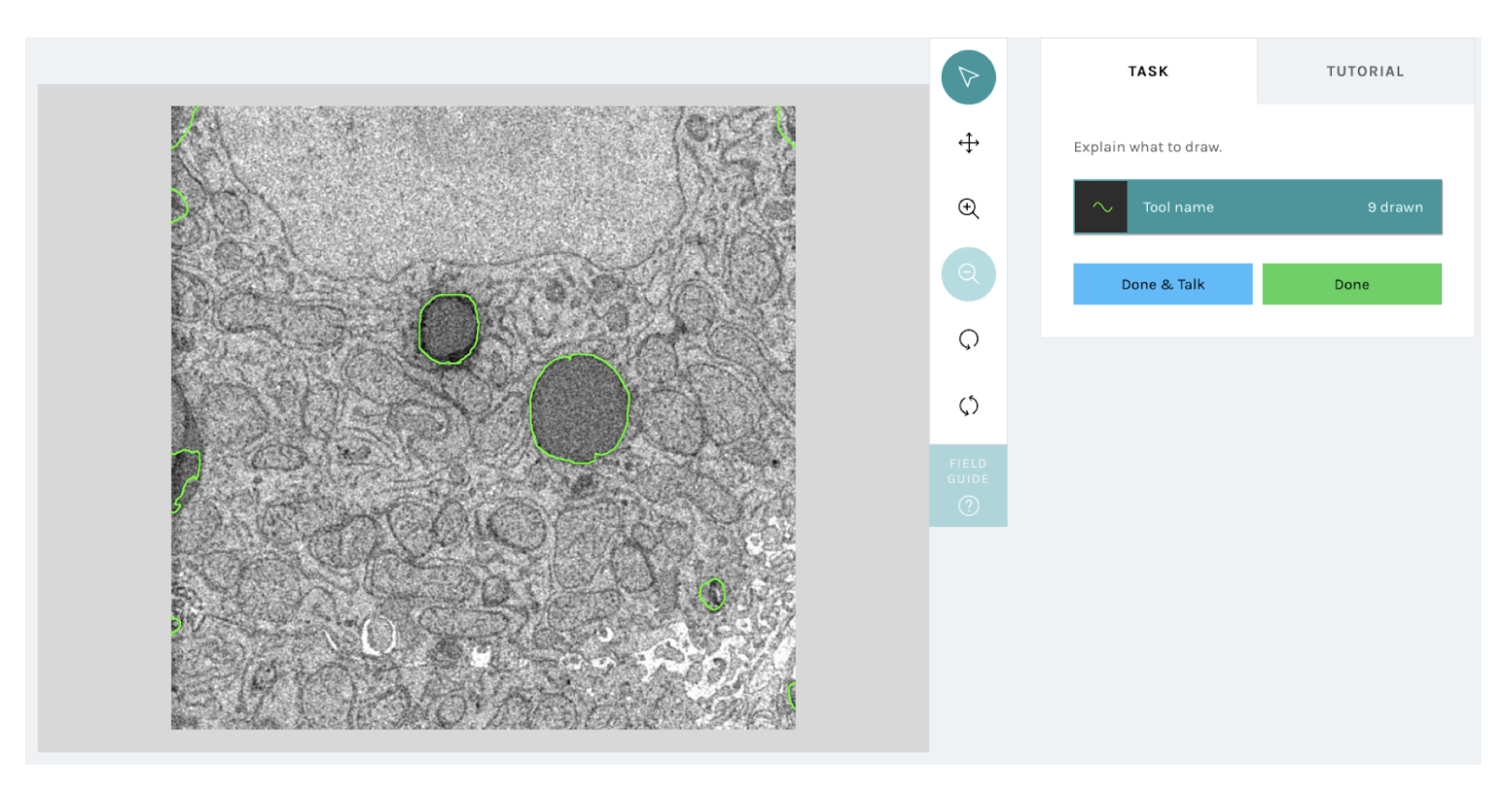
We have built a new infrastructure that will enable researchers to upload machine learning (or any other automated method) based outlines in a compatible format to the freehand line tool such that each subject when viewed by a volunteer on a Zooniverse will be shown the pre-loaded machine outlines, which they can edit using the above newly-added functionality. Once volunteers provide their corrected/edited annotations, their responses will be recorded and used by the research teams for their downstream analyses. The figure below shows an example visualization of what a volunteer would see with the new correct a machine workflow. Note, that the green outlines shown on top of the subject image are directly loaded from a machine model prediction and volunteers will be able to interact with them.
From Fat Droplets to Floating Forests
Finally, the volunteer annotations submitted to these two projects will have an impact far beyond the fat droplets identification in biomedical imaging. Inspired by the flexibility of the PatchGAN model, we also carried out a “Transfer Learning” experiment, where we tested if a model trained to identify fat droplets can be used for a different task of identifying kelp beds in satellite imaging. For this, we used the data from another Zooniverse project called Floating Forests.
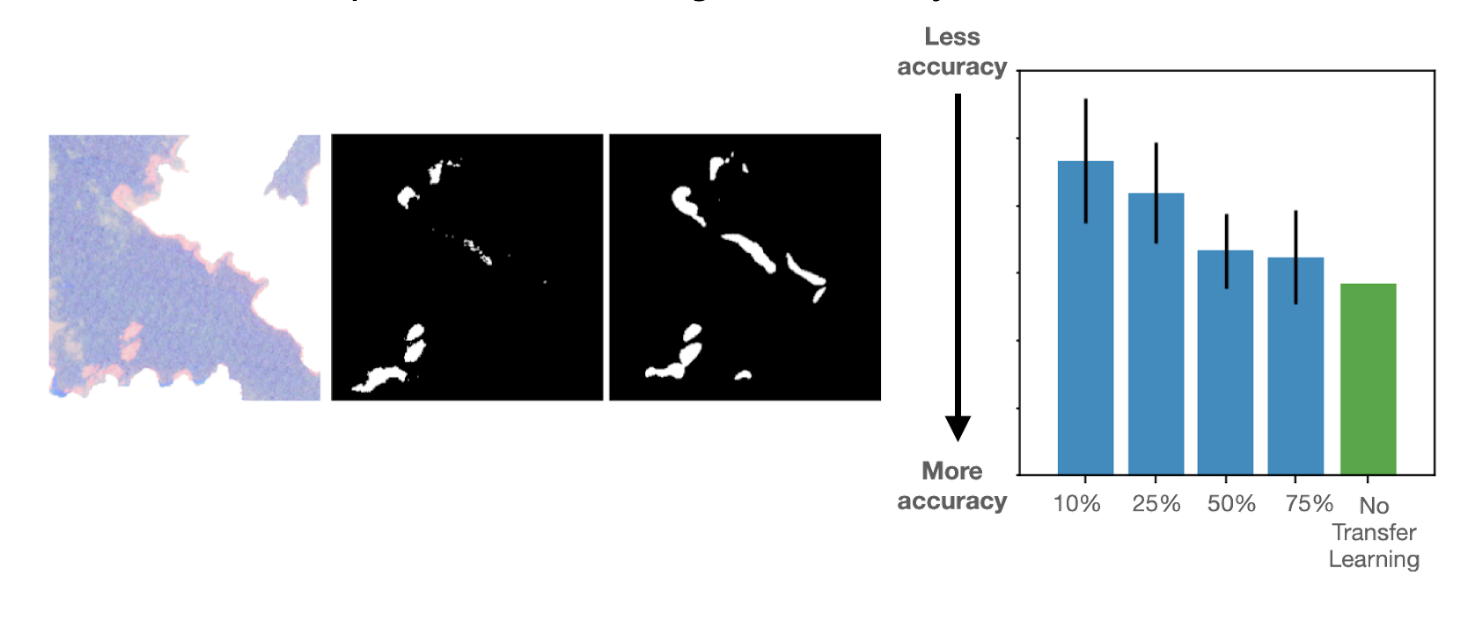
Through this work, we found that our PatchGAN framework readily works to predict the kelp regions. More interestingly, we found that when our pre-trained model to detect fat droplets within Fat Checker project data was used as a starting point to predict the kelp regions, the resultant model achieved very good accuracies with only a small number of training images (~10-25% of the overall data set size). Below is an example subject along with the volunteer annotated kelp regions and corresponding PatchGAN prediction. The bar chart illustrates how the Etch A Cell – Fat Checker based annotations can help reduce the amount of annotations required to achieve a good accuracy.
In summary: THANK YOU!
In summary, with the help of your participation in the Etch A Cell – Fat Checker and Etch A Cell – Fat Checker Round 2 projects, we have made great strides in processing the data and training various successful machine learning frameworks. We have also made a lot of progress in updating the annotation tools and building new infrastructure towards making the best partnership between humans and machines for science. We are looking forward to launching new projects that use this new infrastructure we have built!
This project is part of the Etch A Cell organisation
‘Etch A Cell – Fat Checker’ and ‘Etch A Cell – Fat Checker round 2’ are part of The Etchiverse: a collection of multiple projects to explore different aspects of cell biology. If you’d like to get involved in some of our other projects and learn more about The Etchiverse, you can find the other Etch A Cell projects on our organisation page here.
Thanks for contributing to Etch A Cell! – Fat Checker!