Below is a guest blog post from Dr Philip Fowler, lead researcher on our award-winning biomedical research project Bash the Bug. Read on to find out more about this project and how you can get involved!
– Helen
Our bug-squishing project, BashTheBug, was six months old this month. Since launching on 7th April 2017, over seven thousand Zooniverse volunteers have contributed nearly half a million classifications between them, making 58 classifications per person, on average.
The bugs our volunteers have been bashing are the bacterium responsible for Tuberculosis (TB); ‘Mycobacterium Tuberculosis’. Many people think of TB as a disease of the past, to be found only in the books of Charles Dickens. However, the reality is quite different; TB is now responsible for more deaths each year than HIV/AIDS; in 2015 this disease killed 1.8 million people. To make matters worse, like all other bacterial diseases, TB is evolving resistance to the antibiotics used to treat it. It is this problem that inspired the BashTheBug project, which aims to improve both the diagnosis and treatment of TB.
At the heart of this project is the simple idea that, in order to find out which antibiotics are effective at killing a particular TB strain, we have to try growing that strain in the presence of a range of antibiotics at different doses. If an antibiotic stops the bacterium growing at a dose that can be used safely within the human body, then bingo! that antibiotic can be used to treat that strain. To make doing this simpler, the CRyPTIC project (which is an international consortium of TB research institutions), has designed a 96-well plate which has 14 different anti-TB drugs freeze-dried to the bottom of each well.
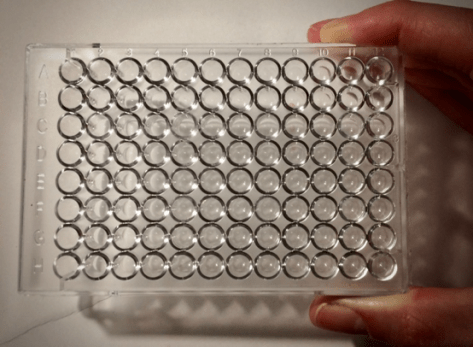
Figure 1. A 96-well microtitre plate
These plates are common in science and are about the size of a large mobile phone. When a patient comes into clinic with TB, a sample of the bacterium they are infected with is taken, grown for a couple of weeks and then some is added to each of the 96 wells. The plate is then incubated for two weeks, and then examined to see which wells have TB growing in them and which do not. As each antibiotic is included on the plate at different doses, it is possible to work out the minimum concentration of antibiotic that stops the bug from growing.
But why are we doing this? Well, the genome of each TB sample will also be sequenced. This will allow us to build two large datasets; one of the mutations in the TB genome and another listing which antibiotics work for each sample (and which do not). Using these two datasets, we will then be able to infer which genetic mutations are responsible for resistance to specific antibiotics. With me still? Good. This will give researchers a large and accurate catalogue that would allow anyone to predict which antibiotics would work on any TB infection, simply by sequencing its genome. This is particularly important for the diagnosis and treatment of TB; currently used approaches are notoriously slow, taking up to eight weeks to identify which antibiotics can be used for effective treatment. If you were a clinician would you want to wait two months before starting your patient on treatment? Of course not.

Figure 2. A photograph of M. tuberculosis that has been growing on a plate for two weeks.
You might scoff at this point and say, pah, using genetics like this in hospitals will never happen. Well it already is. Since March 2017, all routine testing for Tuberculosis in England has been done by sequencing the genome of each sample that is sent to either of the two Public Health England reference laboratories. A report is returned to the clinician in around 9 days. Surprisingly, this costs less than the old, traditional methods for TB diagnosis and treatment. Sequencing TB samples also provides other valuable information, for example, you can compare the genomes of different infections to determine if an outbreak is underway, at no extra cost.
So far, so good. The main challenge to this project though, is size. We will be collecting around 100,000 samples from people with TB from around the world between now and 2020. Every single sample will have its genome sequenced and its susceptibility to different antibiotics tested on our 96-well plates. Each of these plates then need to be looked at, and any errors or inconsistencies in how this huge number of 96 well plates are read could lead to false conclusions about which mutations confer resistance, and which don’t.
This problem is why we need your help! You might not be clinical microbiologists (although a few of you no doubt are!) but there are many, many more of you than we have experienced and trained scientists. In fact, each plate will only be looked at by one, maybe two, scientists, and so it is highly likely that, without the help of volunteers, our final dataset will be riven with differences due to how different people in different labs have read the plates. The inconvenient truth, however much we’d like to think otherwise, is staring at a small white circle and deciding whether there is any M. tuberculosis growing or not is a highly subjective task. Take a look at the strip of wells below – the two wells in the top left have no antibiotic at all so give you an idea of how this strain of TB grows normally.

Figure 3. Is there a dose above which the bacteria doesn’t grow?
In the BashTheBug project, you are asked if there is a dose of antibiotic above which the antibiotic doesn’t grow. If you think there is, you are then asked the number of the first well that doesn’t have any TB growing. For the example image above, I might be cautious and say, well, I can see that there appears to be less and less growth as we go to the right and the dosage increases, but it never entirely goes away; there is a very, very faint dot in well #8. So I’m going to say that actually I think there is bacterial growth in all eight wells. You might be optimistic (or even just in a good mood) and disagree with me and say, yes, but by the time you get to well #6, that dot is so small compared to the growth in the control wells, either the antibiotic is doing its job, or, you know what, I’m not convinced that the dot isn’t some sediment or something else entirely.
There is no correct answer. We are probably both right to some extent; there IS something in well #8, but maybe this antibiotic would still be an effective treatment as it would be able to kill enough of the bacteria for your immune system to then be able to kill off the remainder of the infection. Therefore, the aim of BashTheBug is to identify which antibiotic dose multiple people agreed is the dose above which the bacteria no longer grows. Our result from this project is the consensus we get from showing each image to multiple people. Yes, the volunteers might, on average, take a slightly different view to an experienced clinical microbiologist, but that doesn’t matter as they will, on average, be consistent across all the plates which is vital if we are to uncover which genetic mutations confer resistance to antibiotics.
None of this would be possible without the hard work of all our volunteers. So, if you’ve done any classifications, thank you for all your help. Here’s to another six months, many more classifications, and the first results from the hard work done by the many volunteers who have taken part in the project to date.
Find out more:
- Contribute to the project here
- Read the official BashTheBug blog here
- Follow @BashTheBug on Twitter here
- BashTheBug won the Online Community Award of the NIHR Let’s Get Digital Competition, read more here
Check out other coverage of BashTheBug:
